1 MEAPLQTGMVLGVMIGAGVAVVVTAVLILLVVRRLRVPKTPAPDGPRYRFRKRDKVLFYG
61 RKIMRKVSQSTSSLVDTSVSATSRPRMRKKLKMLNIAKKILRIQKETPTLQRKEPPPAVL
121 EADLTEGDLANSHLPSEVLYMLKNVRVLGHFEKPLFLELCRHMVFQRLGQGDYVFRPGQP
181 DASIYVVQDGLLELCLPGPDGKECVVKEVVPGDSVNSLLSILDVITGHQHPQRTVSARAA
241 RDSTVLRLPVEAFSAVFTKYPESLVRVVQIIMVRLQRVTFLALHNYLGLTNELFSHEIQP
301 LRLFPSPGLPTRTSPVRGSKRMVSTSATDEPRETPGRPPDPTGAPLPGPTGDPVKPTSLE
361 TPSAPLLSRCVSMPGDISGLQGGPRSDFDMAYERGRISVSLQEEASGGSLAAPARTPTQE
421 PREQPAGACEYSYCEDESATGGCPFGPYQGRQTSSIFEAAKQELAKLMRIEDPSLLNSRV
481 LLHHAKAGTIIARQGDQDVSLHFVLWGCLHVYQRMIDKAEDVCLFVAQPGELVGQLAVLT
541 GEPLIFTLRAQRDCTFLRISKSDFYEIMRAQPSVVLSAAHTVAARMSPFVRQMDFAIDWT
601 AVEAGRALYRQGDRSDCTYIVLNGRLRSVIQRGSGKKELVGEYGRGDLIGVVEALTRQPR
661 ATTVHAVRDTELAKLPEGTLGHIKRRYPQVVTRLIHLLSQKILGNLQQLQGPFPAGSGLG
721 VPPHSELTNPASNLATVAILPVCAEVPMVAFTLELQHALQAIGPTLLLNSDIIRARLGAS
781 ALDSIQEFRLSGWLAQQEDAHRIVLYQTDASLTPWTVRCLRQADCILIVGLGDQEPTLGQ
841 LEQMLENTAVRALKQLVLLHREEGAGPTRTVEWLNMRSWCSGHLHLRCPRRLFSRRSPAK
901 LHELYEKVFSRRADRHSDFSRLARVLTGNTIALVLGGGGARGCSHIGVLKALEEAGVPVD
961 LVGGTSIGSFIGALYAEERSASRTKQRAREWAKSMTSVLEPVLDLTYPVTSMFTGSAFNR
1021 SIHRVFQDKQIEDLWLPYFNVTTDITASAMRVHKDGSLWRYVRASMTLSGYLPPLCDPKD
1081 GHLLMDGGYINNLPADIARSMGAKTVIAIDVGSQDETDLSTYGDSLSGWWLLWKRLNPWA
1142 DKVKVPDMAEIQSRLAYVSCVRQLEVVKSSSYCEYLRPPIDCFKTMDFGKFDQIYDVGYQ
1201 YGKAVFGGWSRGNVIEKMLTDRRSTDLNESRRADVLAFPSSGFTDLAEIVSRIEPPTSYV
1261 SDGCADGEESDCLTEYEEDAGPDCSRDEGGSPEGASPSTASEMEEEKSILRQRRCLPQEP
1321 PGSATDA
Neuropathy Target Esterase (NTE) and the Cholinergic System
What is NTE?
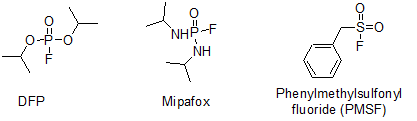
Neuropathy target esterase (NTE) is an integral membrane protein that is anchored to the endoplasmic reticulum (ER) and is known to be involved in lipid hydrolysis. Furthermore, NTE is known to be the cellular target for a class of organophosphorus compounds (OPs) termed neuropathic OPs that can, following exposure to a sufficiently toxic dose, initiate a delayed axonopathy. This condition, known as OP-induced delayed neurotoxicity (OPIDN), is understood follow inexorably after the inhibition of >70% of neuronal NTE by neuropathic OPs (such as diisopropyl phosphorofluoridate (DFP) and diisopropyldiamidophosphorofluoridate (Mipafox)), followed by an 'aging' reaction involving the OP adduct. Aging is a post-inhibitory reaction wherein the NTE-OP conjugate gains a net negative charge. In most cases, this occurs via the net loss of an alkyl (R) group from the OP adduct bound to the active site serine residue (Ser966) of NTE. The end result of the aging reaction is a permanently inhibited enzyme that cannot be reactivated via the use of strong nucleophiles like fluoride.
Structure of NTE
While the tertiary structure of NTE is currently unknown, it is known to be a multi-domain serine hydrolase consisting of an N-terminal transmembrane domain (TMD), three cyclic nucleotide binding domains and a C-terminal catalytic domain that is responsible for the lipid hydrolysis activity of the enzyme (Fig. 1). The catalytic domain is known to be similar to the plant protein Patatin and we have shown its catalytic center to consist of a Ser966-Asp1086 catalytic dyad, similar to that found in patatin and cytosolic phospholipase A2. Indeed, it is now recognized that NTE is a member of a broader family of phospholipases termed the patatin-like phospholipase A2 (PNPLA) family of which, NTE is the sixth member (hence its designation as PNPLA6).
Primary amino acid sequence of NTE (isoform 2, which was the first isorform to be sequenced):
While the tertiary structure of NTE has yet to be elucidated, the structure of Patatin in complex with aging and non-aging OP compounds has been reported (Wijeyesakere et al., 2014).